FREEDOM!
At around 2130 last evening, I noticed that the ship was no longer following the typical pattern of the last six days (reverse/forward/ram/repeat). Instead, we were moving forward – and we were not stopping. After 6+ days in the heavily packed, snow covered ice, we reached a point where the snow and ice combination was small enough to allow us to break through and keep going. I went to the bridge to get a bird's eye view of the ice edge. Without much fanfare, we left the pack ice at 2240. It was a bitter-sweet moment for me. It will most likely be the last time I see the mountains of Antarctica and the pack ice (that's the bitter part). The sweet part has to do with the science. Our departure from the ice allows us to squeeze in one more day of science before we must depart for the Drake Passage and ultimately Chile. The science team has decided on a deep trace metal cast (3500m/10500ft) followed by water collection. I will explain more about this in tomorrow's journal.
Guest Author
Kris Gomes from the University of Rhode Island expressed interest in writing one of the journals. With just a few days left in the research cruise, I thought it might be great if Kris could explain a word that has been tossed around for the past few weeks...metatranscriptomes. I have seen large carboys of water being carted around the ship from deck-board incubators to walk-in coolers, but I really was not sure why these large quantities of water were necessary. Thanks to Kris' explanation, I have a better understanding of this important analysis technique. The following is what Kris had to say about metatranscriptomes.
 The flow-through incubators bring sea water through pumps to the incubators. This allows the bottles to receive ambient (natural) temperature and light during incubation.
The flow-through incubators bring sea water through pumps to the incubators. This allows the bottles to receive ambient (natural) temperature and light during incubation.
 The 20L carboys remained in the incubators for 12 days. You may notice an Antarctic krill swimming in the incubator. The krill traveled into the system as sea water was pumped from the surroundings through the pipes of the incubator system.
The 20L carboys remained in the incubators for 12 days. You may notice an Antarctic krill swimming in the incubator. The krill traveled into the system as sea water was pumped from the surroundings through the pipes of the incubator system.
The Central Dogma
Metatranscriptomes are an important tool biologists can use to understand how communities of cells in an environment, such as diatoms and bacteria in the Southern Ocean, respond to their environment and to each other. Before we talk about what metatranscriptomes are its important to understand the relationship between DNA, RNA, and proteins. In a previous journal, we discussed the importance of DNA and RNA molecules and their ability to convey genetic information. This relationship is a part of what is referred to as the central dogma, the foundation of molecular biology, which describes the flow of genetic information from DNA to RNA and finally to the production of proteins.
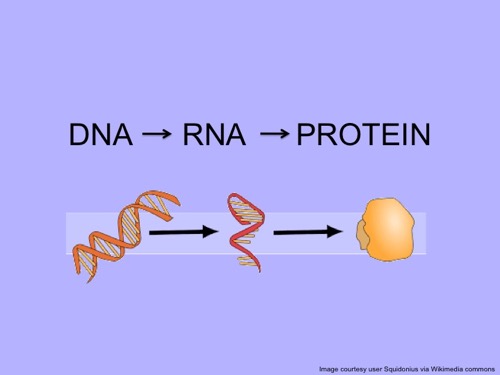 Diagram showing the flow of genetic information from the DNA inside the cell's nucleus to the production of proteins for structural and chemical functions.
Diagram showing the flow of genetic information from the DNA inside the cell's nucleus to the production of proteins for structural and chemical functions.
In a cell, DNA serves as the blueprint of all the proteins that an organism is capable of expressing (making). During the process of transcription, or RNA synthesis, the genetic code for specific proteins are copied into RNA transcripts, acting as “instructions” telling the cell which proteins should be made. These “instructions” are then read by cell parts called ribosomes and used to produce proteins. This process is referred to as translation. The proteins act as the “functional tools” of the cell and/or the organism. These proteins can act as enzymes (like the lactase that breaks down the lactose sugars in the milk that you have with breakfast), structural proteins (like the keratin protein that makes up your hair and nails), etc. Cells can produce different proteins throughout their lives depending on their role inside the organism and/or changes in their environment.
What Is a Metatranscriptome and What Can It Tell Us?
Metatranscriptomes refer to the pool of all RNA transcripts that are being produced by the organisms within a specific community at a specific time. They can be thought of as genetic snapshots of what a community is doing and how they are responding to their environment. In other words, what RNA (and eventually what proteins) are the cells creating at a given moment in time.
Under changing conditions cells are able to transcribe or copy different RNA transcripts and in different amounts in response to their physiological needs or environmental stresses. This change in the number of specific RNA transcripts produced is referred to as differential expression. As we discussed, RNA serves as the “instructions” to the cell of what proteins should be synthesized. Changes in the amounts and types of RNA transcripts produced result in a corresponding change in protein synthesis. The differential expression of RNA transcripts and the resulting proteins allows for an organism to physiologically change in response to differences in their environment, such as the differing nutrient levels being tested in our incubation experiments.
By sequencing the large pool of transcripts collected from these experiments we are able to read and begin to identify the sequence of RNA transcripts. These sequences give us an idea of the proteins being made by the diatoms, or other studied cells. We can begin to understand how the organisms in the community are responding to the different conditions being tested in our incubation experiments. Put simply: do diatoms produce different proteins in response to the different nutrients in each incubation group? This information allows us to understand not only how single organisms respond to the different conditions being tested but also how the community as a whole responds and interacts with each other.
Sample Collection
Sequencing a metatranscriptome requires a large amount of RNA, which means the science team must filter large quantities of water to collect large amounts of biomass. To get the amount of biomass required, the metatranscriptome experiments are setup in large 20 liter carboys, allowing for a greater number of cells to grow during the course of the experiment. Just like the water from the other incubation experiments the metatranscriptome bottles are filtered down to collect all the biomass. The filters are flash frozen in liquid nitrogen and brought back to lab. Once at the lab the pool of RNA from the cells will be extracted and sent out for sequencing, so that we can begin to make comparisons of how the communities collected respond to the different experimental conditions.
 The filtration set-up for four metatranscriptome sampling bottles (clear carboys on counter). The water from the carboys is pumped through the large filters handing into the carboys on the floor of the cold room. Large quantities of water are filtered for metatranscriptome analysis.
The filtration set-up for four metatranscriptome sampling bottles (clear carboys on counter). The water from the carboys is pumped through the large filters handing into the carboys on the floor of the cold room. Large quantities of water are filtered for metatranscriptome analysis.
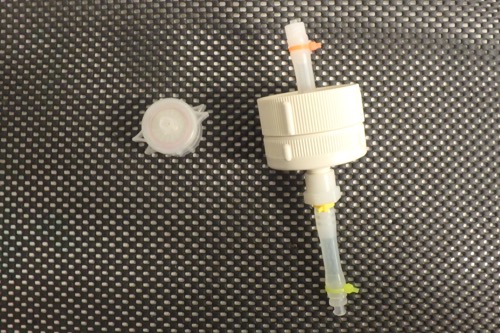 The filter on the left is the 25mm (diameter) filter rig used to extract biomass for RNA and DNA sampling. The filter on the right is the larger (47mm diameter) filter rig used for metatranscriptomes. Both house 3 micron filters in order to catch diatoms and other biomass, but one will hold much more than the other.
The filter on the left is the 25mm (diameter) filter rig used to extract biomass for RNA and DNA sampling. The filter on the right is the larger (47mm diameter) filter rig used for metatranscriptomes. Both house 3 micron filters in order to catch diatoms and other biomass, but one will hold much more than the other.

Comments