Last week I traveled to Corvallis, Oregon to visit Dr. Byron Crump and his lab at Oregon State University. We planned the final dates and details of our summer trip and I learned so much about the research that I will be doing with him.
 Pipetting is one the skills needed in Dr. Crump's lab. Photo by Dr. Crump
Pipetting is one the skills needed in Dr. Crump's lab. Photo by Dr. Crump
Overall description of research that I will be helping with:
What research occurs in the field:
We are traveling to 40 sites around our Toolik Field station in the Arctic tundra watershed in the Northern Part of Alaska.
The research we are doing belongs in the field of microbial biogeography, which is a field that studies how microbes are distributed across a landscape. We are researching what is controlling their diversity and their growth rate.
When we go to these sites we will be thinking about what is affectionately known as meta-communities (meaning lots of different communities that interact through dispersal), it is our job to collect water in the different places where it flows.
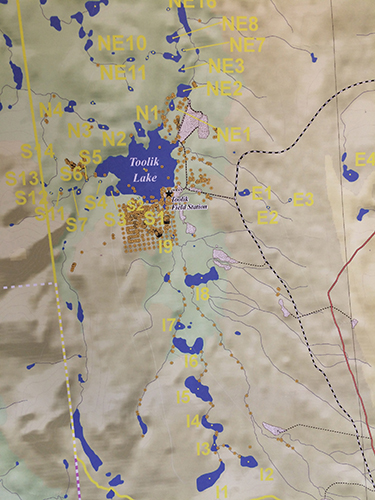 This is a map of Toolik lake and some of the local sites in the watershed that we shall be visiting in June. Photo by DJ Kast
This is a map of Toolik lake and some of the local sites in the watershed that we shall be visiting in June. Photo by DJ Kast
Within Dr. Crump and Dr. Kling’s study we are starting at a high point of a watershed where the rain falls into the soil. We follow the water flow down the watershed through streams, lakes and ponds to map the ecological function that microbes have. The landscape is a seed bank of diversity that sets the scene on the slope through the watershed. Some of the sites are only accessible by helicopter! I am so looking forward to flying in a helicopter for the first time.
Cool fact- Soil samples that are collected will have a lot of the bacteria on the entire planet in terms of diversity. Some are more rare then others but they are all mostly there.
DNA is extracted from all of these water samples and sent to the lab at OSU in freezing temperatures.
 This is a tray of samples filled with water sample DNA of each of the different sites. Photo by DJ Kast
This is a tray of samples filled with water sample DNA of each of the different sites. Photo by DJ Kast
Post processing in Dr. Crump’s lab:
When the DNA extractions from the various sites arrive, all of the DNA in these samples are not only copied but also amplified through PCR (polymerase chain reaction), which means they have a lot more DNA to work with- like millions to billions of copies.
After all the DNA has been amplified, the samples are sent over to a DNA sequencer that spurts out billions of nucleotides which then need to be sorted for each respective species of organism.
The lab and many microbiologists use 16s rRNA genes to identify bacteria. The 16S rRNA gene is a section of prokaryotic DNA found in all bacteria and archaea. This gene codes for an rRNA, and this rRNA in turn makes up part of the ribosome. The first 'r' in rRNA stands for ribosomal. When they look at bacteria in lakes there is a lot of discovery and new organisms are found all the time with their DNA presence within a sample of a watershed.
 Dr. Crump reviewing the PCR strength of his DNA extractions. Photo by DJ Kast
Dr. Crump reviewing the PCR strength of his DNA extractions. Photo by DJ Kast
“Bacteria is a biological frontier that has a lot of discovery. Half of the genes that we find in our environmental surveys are genes that we don’t know the function of.” Dr. Crump says ”One of the reasons I came to Oregon State University (OSU) was because of their amazing DNA sequencing center. It’s a hub for experts in bioinformatics; I knew my research would thrive here.“
This is just a glimpse of the research and post-processing of samples that will be happening in the Arctic Field station of Toolik.


Comments